-Search query
-Search result
Showing 1 - 50 of 81 items for (author: gan & sy)

EMDB-19406: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L

EMDB-19407: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L

PDB-8rox: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L

PDB-8roy: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24
Method: single particle / : Shilliday F, Lucas SCC, Richter M, Michaelides IN, Fusani L
EMDB-28138: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
EMDB-28141: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
EMDB-28142: 
3D Reconstruction of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY

EMDB-42804: 
Structure of nucleotide-free Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D

EMDB-42806: 
Structure of AMP-PNP-bound Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D

EMDB-42807: 
Structure of ADP-bound and phosphorylated Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D

PDB-8uyf: 
Structure of nucleotide-free Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D

PDB-8uyh: 
Structure of AMP-PNP-bound Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D

PDB-8uyi: 
Structure of ADP-bound and phosphorylated Pediculus humanus (Ph) PINK1 dimer
Method: single particle / : Gan ZY, Kirk NS, Leis A, Komander D

EMDB-36887: 
Structure of GPR34-Gi complex
Method: single particle / : Wong TS, Zeng ZC, Xiong TT, Gan SY, He GD, Du Y

EMDB-28125: 
Plasmodium falciparum merozoites apical 2-ring units
Method: subtomogram averaging / : Sun SY, Pintilie GD

EMDB-28126: 
Toxoplasma apical rings
Method: subtomogram averaging / : Sun SY, Pintilie GD

EMDB-28910: 
Glycan-Base ConC Env Trimer
Method: single particle / : Olia AS, Kwong PD

EMDB-27630: 
Structure of the PEAK3/14-3-3 complex
Method: single particle / : Torosyan H, Paul M, Jura N, Verba KA

EMDB-27684: 
Structure of the PEAK3 pseudokinase homodimer
Method: single particle / : Torosyan H, Paul M, Jura N, Verba KA

PDB-8dp5: 
Structure of the PEAK3/14-3-3 complex
Method: single particle / : Torosyan H, Paul M, Jura N, Verba KA

PDB-8ds6: 
Structure of the PEAK3 pseudokinase homodimer
Method: single particle / : Torosyan H, Paul M, Jura N, Verba KA

EMDB-29396: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD
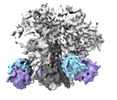
EMDB-29836: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-29880: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29881: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29882: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)
Method: single particle / : Changela A, Gorman J, Kwong PD

EMDB-29905: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8fr6: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD

PDB-8g85: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8g9w: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8g9x: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8g9y: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)
Method: single particle / : Changela A, Gorman J, Kwong PD

PDB-8gas: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-28139: 
Toxoplasma gondii apical complex (ionophore stimulated)
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY, Egan ES, Chiu W, Boothroyd JC

EMDB-28140: 
Toxoplasma gondii apical complex (non-stimulated)
Method: electron tomography / : Segev-Zarko L, Sun SY, Chiu W, Boothroyd JC

EMDB-13776: 
Structure of formaldehyde cross-linked SARS-CoV-2 S glycoprotein
Method: single particle / : Sulbaran G, Effantin G, Schoehn G, Weissenhorn W

PDB-7q1z: 
Structure of formaldehyde cross-linked SARS-CoV-2 S glycoprotein
Method: single particle / : Sulbaran G, Effantin G, Schoehn G, Weissenhorn W

EMDB-25677: 
Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1
Method: single particle / : Gan ZY, Leis A, Dewson G, Glukhova A, Komander D

EMDB-25678: 
Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with kinked alpha-C helix in chain B
Method: single particle / : Gan ZY, Leis A, Dewson G, Glukhova A, Komander D

EMDB-25679: 
Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with extended alpha-C helix in chain B
Method: single particle / : Gan ZY, Leis A, Dewson G, Glukhova A, Komander D

EMDB-25680: 
Structure of dodecameric unphosphorylated Pediculus humanus (Ph) PINK1 D357A mutant
Method: single particle / : Gan ZY, Leis A

EMDB-25681: 
Structure of dimeric unphosphorylated Pediculus humanus (Ph) PINK1 D357A mutant
Method: single particle / : Gan ZY, Leis A

PDB-7t4k: 
Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with kinked alpha-C helix in chain B
Method: single particle / : Gan ZY, Leis A, Dewson G, Glukhova A, Komander D

PDB-7t4l: 
Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with extended alpha-C helix in chain B
Method: single particle / : Gan ZY, Leis A, Dewson G, Glukhova A, Komander D

PDB-7t4m: 
Structure of dodecameric unphosphorylated Pediculus humanus (Ph) PINK1 D357A mutant
Method: single particle / : Gan ZY, Leis A, Dewson G, Glukhova A, Komander D

PDB-7t4n: 
Structure of dimeric unphosphorylated Pediculus humanus (Ph) PINK1 D357A mutant
Method: single particle / : Gan ZY, Leis A, Dewson G, Glukhova A, Komander D

EMDB-30883: 
Cryo-EM structure of Dengue virus serotype 1 strain WestPac 74 in complex with human antibody 1C19 Fab at 37 deg C (Class 1 particle)
Method: single particle / : Fibriansah G, Ng TS

EMDB-30884: 
Cryo-EM structure of Dengue virus serotype 1 strain WestPac 74 in complex with human antibody 1C19 Fab at 37 deg C (Class 2 particle)
Method: single particle / : Fibriansah G, Ng TS

EMDB-30885: 
Cryo-EM map of Dengue virus serotype 2 strain PVP94/07 in complex with human antibody 1C19 Fab at 37 deg C
Method: single particle / : Fibriansah G, Ng TS, Kostyuchenko VA, Shi J, Lok SM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
